PHYS/BIOL 3250/4814 is an undergraduate level course taught by Professor Daniel I. Goldman in the School of Physics at the Georgia Institute of
Technology. This course is built around modules on topics from protein folding to cell motility to neuromechanics to dynamical systems in ecology and evolution; each module
is taught by Dr. Goldman using materials from PoLS
(physics of living systems) colleagues.
In each module:
1st week: Lecture based around materials from colleagues, textbooks, and literature, on a different topic every other week; curriculum mirroring BIOL 1510.
On Thursday of week one in each module, students “meet the expert”; a colleague visits and provides input in his/her area of expertise.
2nd week: Hands on-lab, “Micro-labs” (one week experiences, every other week) use low cost sensors (e.g. in smartphones) and open source computational packages
to provide hands-on introduction to a particular area. Students work in teams of 2 (4-5 groups). The goal is to get a scientific result comparing experiment and theory/model by the end of the week. Micro-lab pedagogy is built off Dr. Goldman’s experience teaching in the International Hands-On Schools and GT-FIRE experience.
There is no textbook for this class; source material is pulled from open source texts (e.g. http://openstaxcollege.org/), papers, and expertise from guest lecturers.

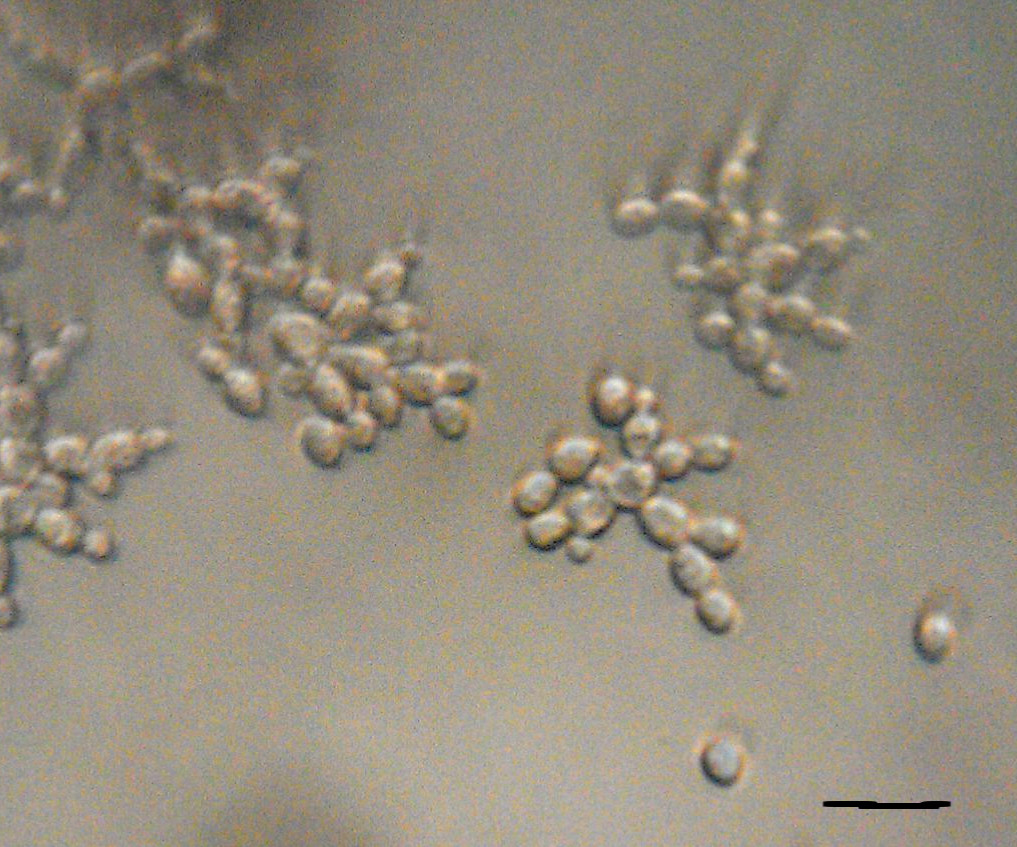

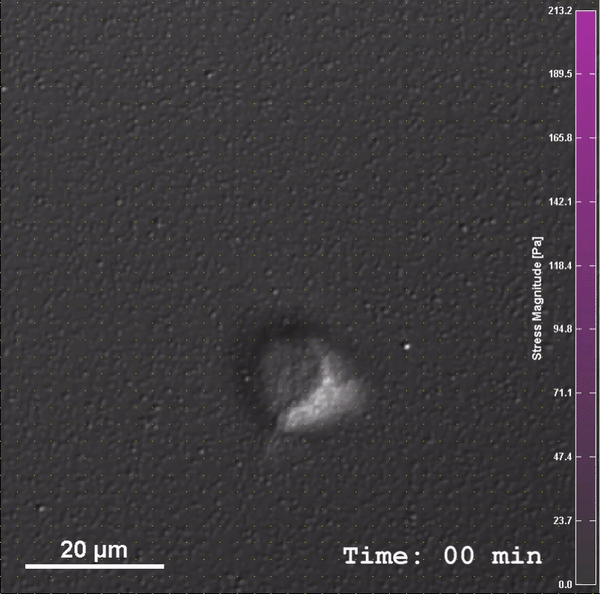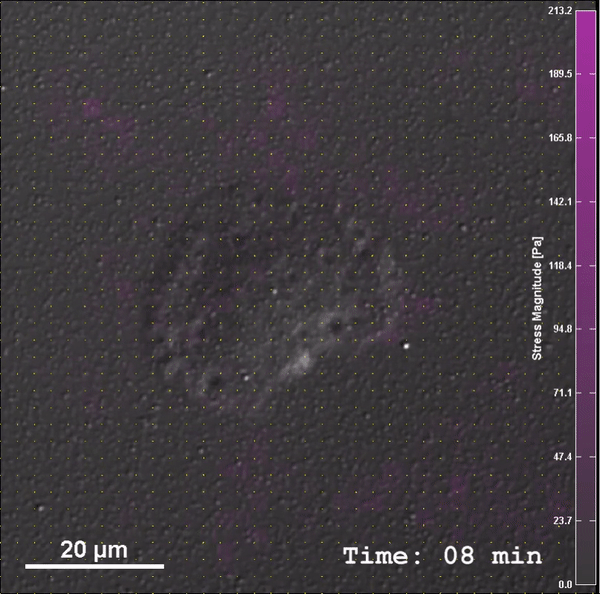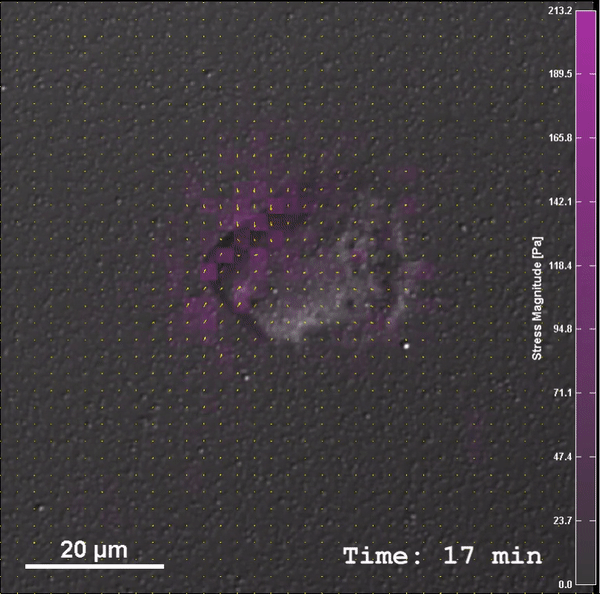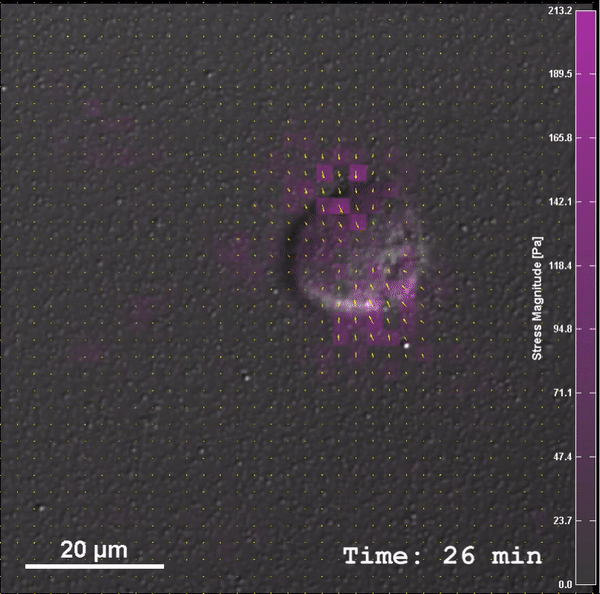

Through work on modules, students will be introduced to PoLS topics and gain confidence and experience with relevant multi-scale biophysical experimental, computational, and theoretical techniques, as well as be introduced to a common language of nonlinear dynamics. After completing this class, students will be able to: (1) Construct low-cost laboratory equipment to investigate fundamental biophysical processes, (2) Integrate measurement, mechanism, and modeling in the study of these systems, and (3) Analyze biophysical systems with computational and theoretical tools (i.e. nonlinear dynamics, molecular dynamics simulations, and statistical mechanics).